This Python package is designed match short peptide sequences detected via Mass Spectroscopy to a FASTA file then produce alignment outputs in various formats. An input file format would be:
Peptide F145I/Dd2Dd2 Mass_Spec_Mode
VG;GV 3.493 POS
PA 2.454 POS
SP 4.701 NEG
A collaborator asked me to align short peptides from a Mass Spec experiment to a sequence, then show him an image that displays in an easy-to-see format where does each peptide align and which regions are not covered.
For example, when they had a series of short fragments like:
VL LS LSP LSPAD PA NVKAA NVK VKA AA
And an origin sequence of:
VLSPADKTNVKAAWGK
They wanted to see it matched in a way that lets them see which region is well covered and which region is not covered. I came up with the following visualization that also displays individual, non-overlapping alignments with a packing heuristic:
VLSPADKTNVKAAWG
**
VL PA NVKAA
LS NVK
LSP VKA
LSPAD AA
The * above indicates a region that is not covered. In addition they wanted to display different peptides with colors as well.
I was unable to locate a tool that has a similar functionality, hence I wrote this package.
pip install pepaln
python -m pepaln -m fragments.txt -r reference.fa
Generates the files called output.gff, output.txt and output.pdf
The input consists of a tab delimited format with at least three columns:
Peptide F145I/Dd2Dd2 Mass_Spec_Mode
VG;GV 3.493 POS
PA 2.454 POS
SP 4.701 NEG
Where:
- The first column lists the peptide sequence (multiple sequences may be separated with a semicolon
;). - The second column lists a value
- The third column indicates the ionization mode
The reference fasta file may contain more than one target sequence.
>ha
VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPN
>hb
VHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAVMGNPKVKAHGKKVLGAFSDGLAHL
A separate visualization will be generated for each reference sequence.
The tool will generate outputs in three formats TXT, GFF as well as PDF formats. The default filenames are
output.txt,output.gff,output.pdf
You may override each.
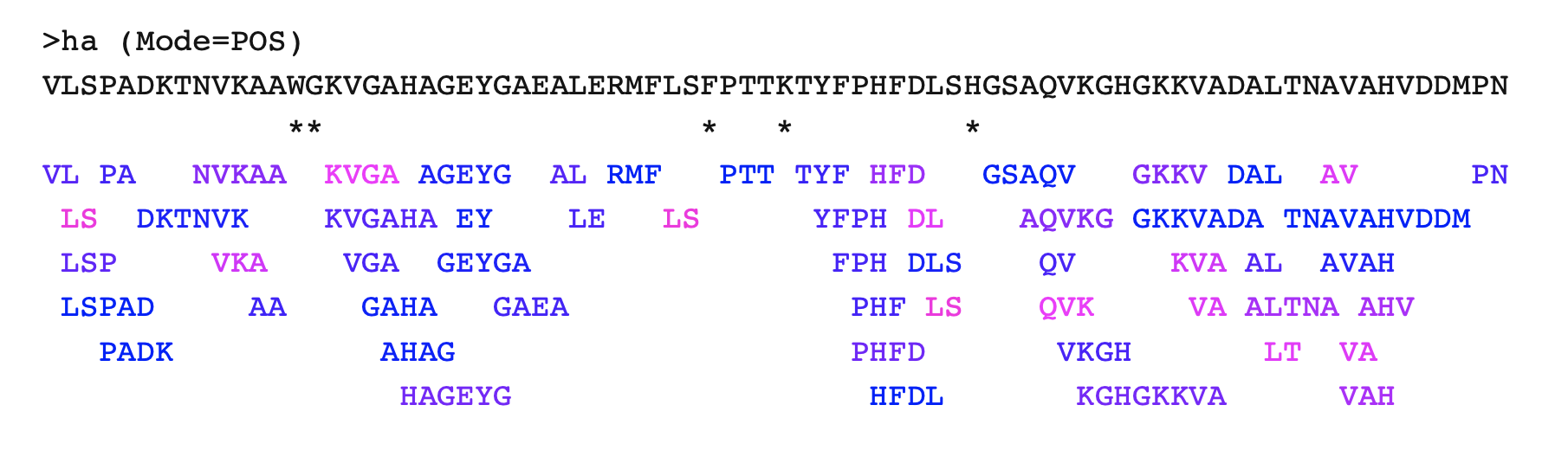
>ha (Mode=POS)
VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPN
** * * *
VL PA NVKAA KVGA AGEYG AL RMF PTT TYF HFD GSAQV GKKV DAL AV PN
LS DKTNVK KVGAHA EY LE LS YFPH DL AQVKG GKKVADA TNAVAHVDDM
LSP VKA VGA GEYGA FPH DLS QV KVA AL AVAH
LSPAD AA GAHA GAEA PHF LS QVK VA ALTNA AHV
PADK AHAG PHFD VKGH LT VA
HAGEYG HFDL KGHGKKVA VAH
The peptides are colored by their value field:
ha VL . 1 2 . 2.433 . Mode=POS
ha LS . 2 3 . 4.806 . Mode=POS
ha LSP . 2 4 . 2.522 . Mode=POS
ha LSPAD . 2 6 . 1.613 . Mode=POS
ha PA . 4 5 . 2.2 . Mode=POS
ha PADK . 4 7 . 1.548 . Mode=POS
ha DKTNVK . 6 11 . 1.845 . Mode=POS
ha NVKAA . 9 13 . 3.012 . Mode=POS
ha VKA . 10 12 . 3.986 . Mode=POS
...
$ python -m pepaln
usage: __main__.py [-h] [-m MASS] [-r REF] [-p output.pdf] [-t output.txt]
[-g output.gff]
optional arguments:
-h, --help show this help message and exit
-m MASS, --mass MASS Mass-spec result file containing peptide sequences.
-r REF, --ref REF Reference file to match the peptides against.
-p output.pdf, --pdf output.pdf
Output file for pdf file
-t output.txt, --txt output.txt
Output file for text alignments
-g output.gff, --gff output.gff
Output file as GFF data