Trying to use Apple's Create ML for Polyps Detection. The result was not impressive though but kind ok for just trying GUI based ML training suite for weekend fun. Thinking of using regular python script with TensorFlow etc for my next experiments.
Somewhat useful tool in this project is convert.py script which you can convert hyper-kvasir data bounding box json into CreateML annotations.json format. It also move files, and generate index.html to preview the bounding box on your browser.
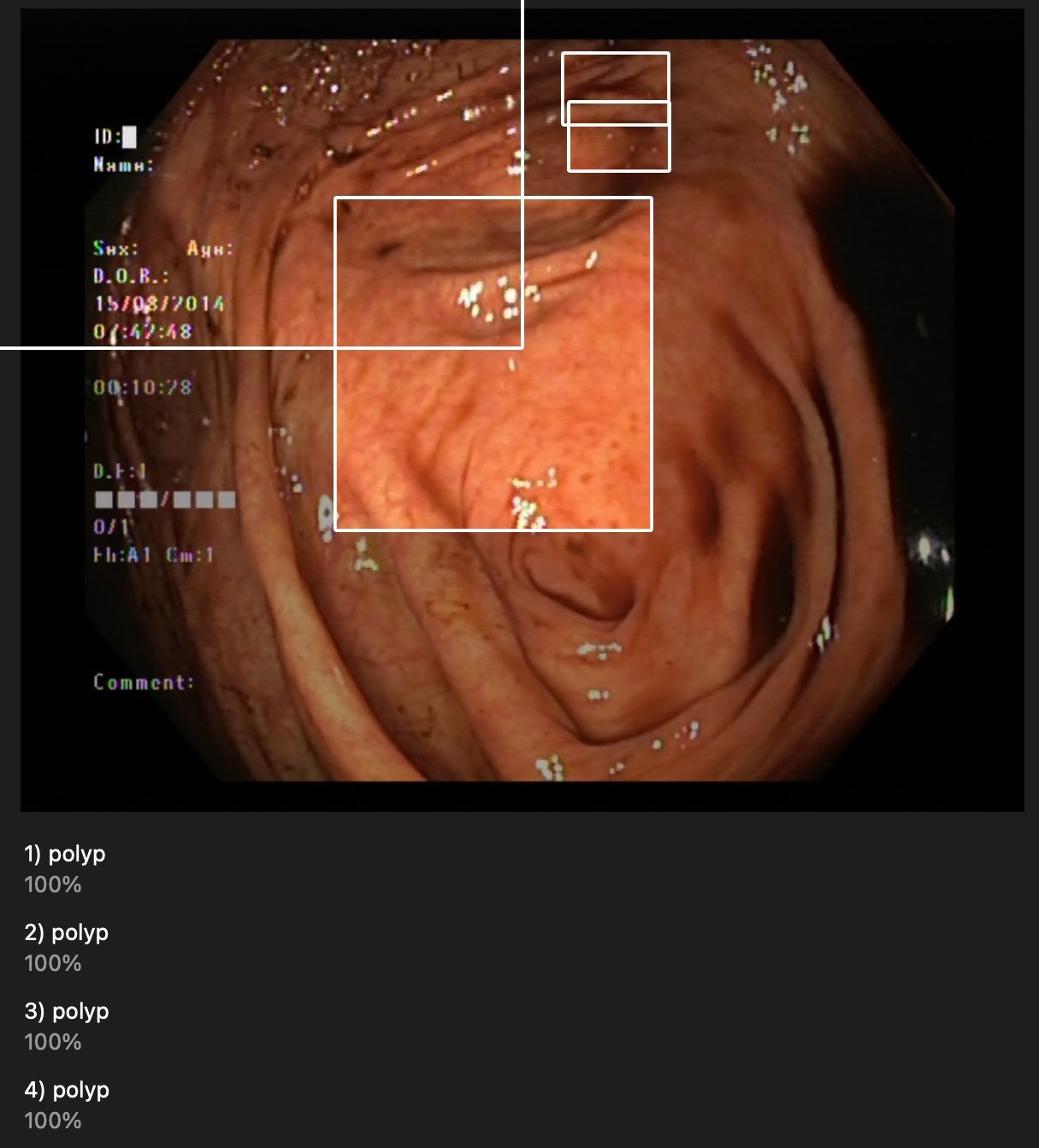
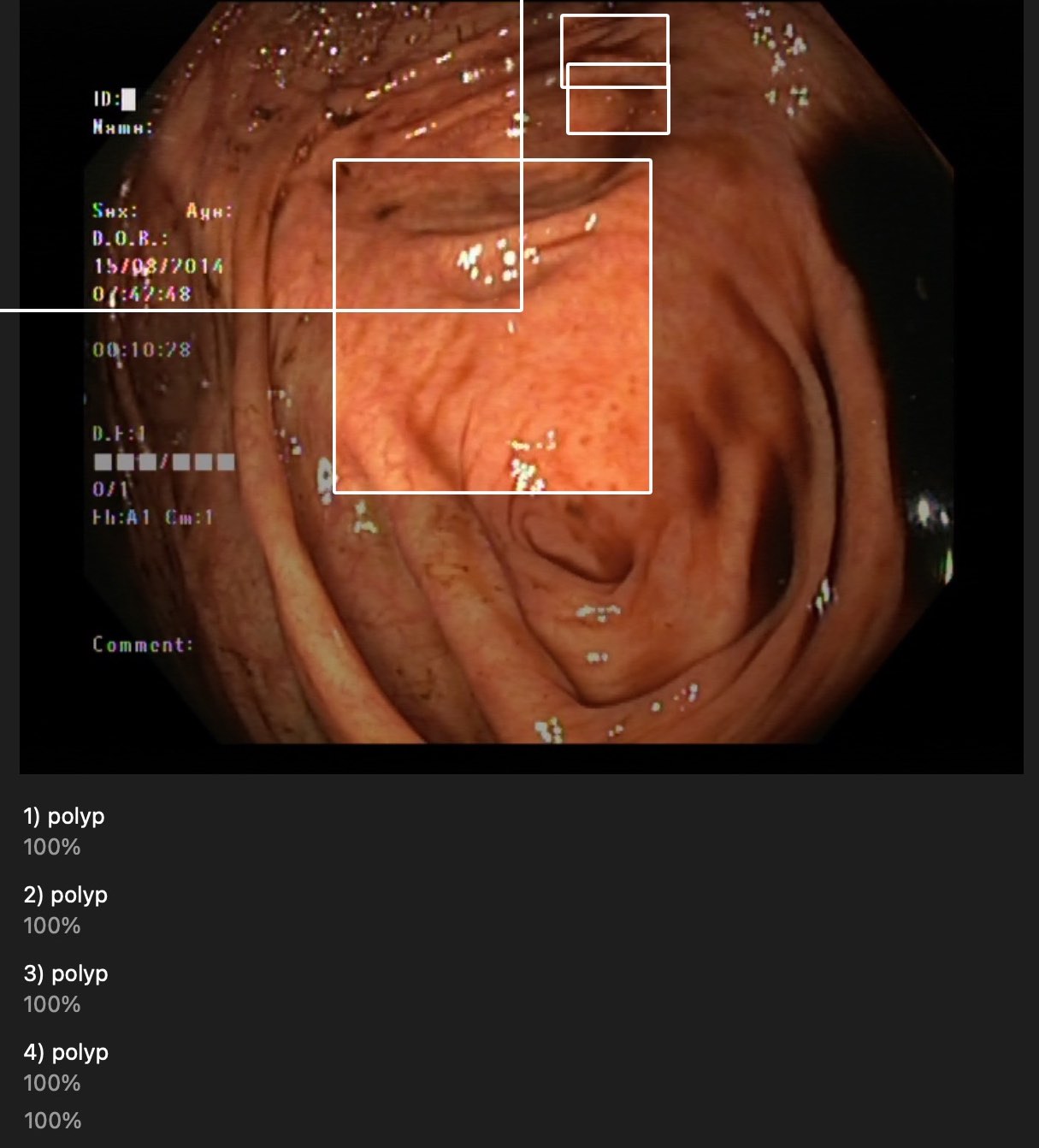
They are near polyps but most of them are not surrounding correctly.
It detected in non-polyps pictures or even on chicken. duh
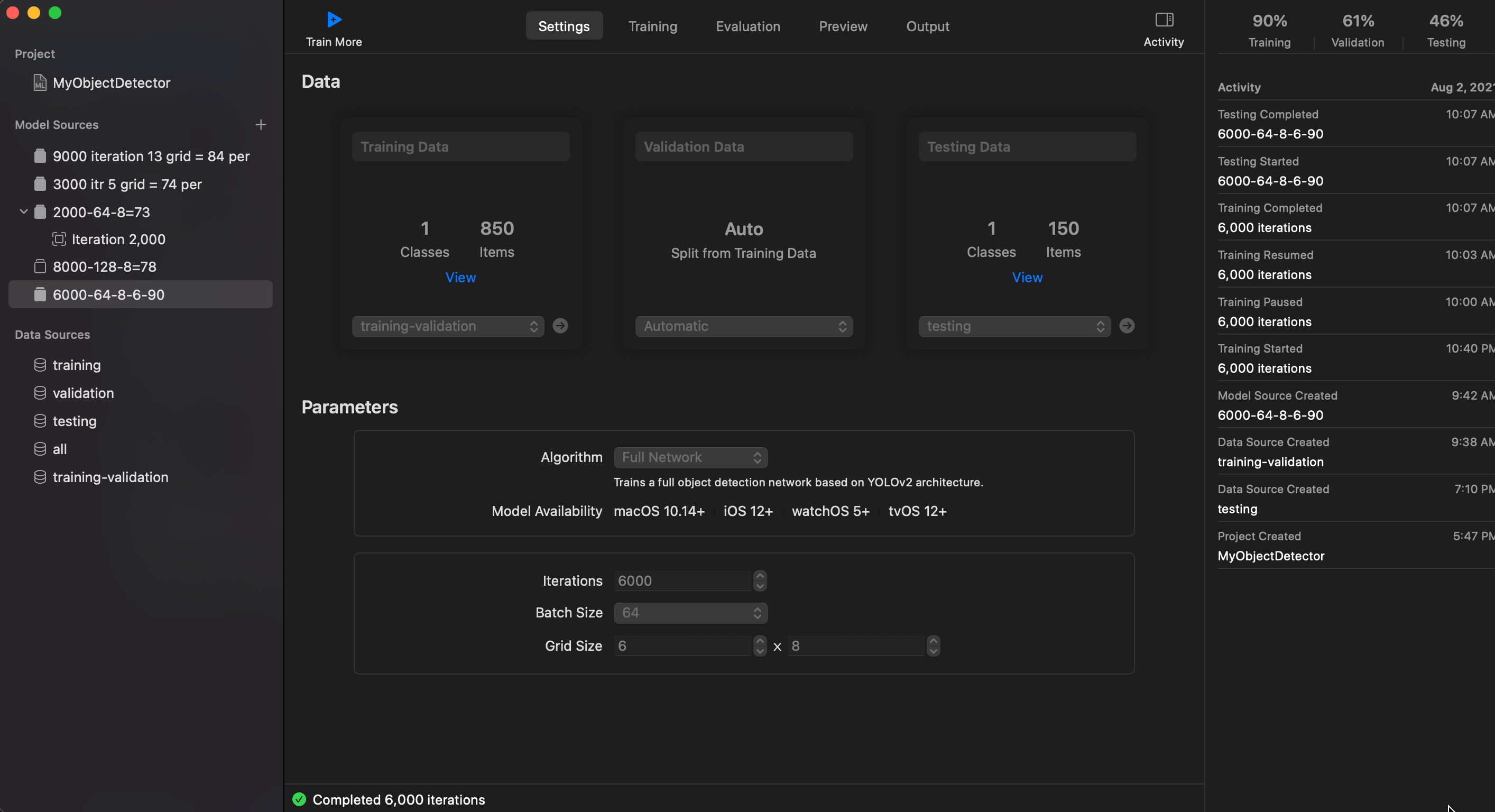
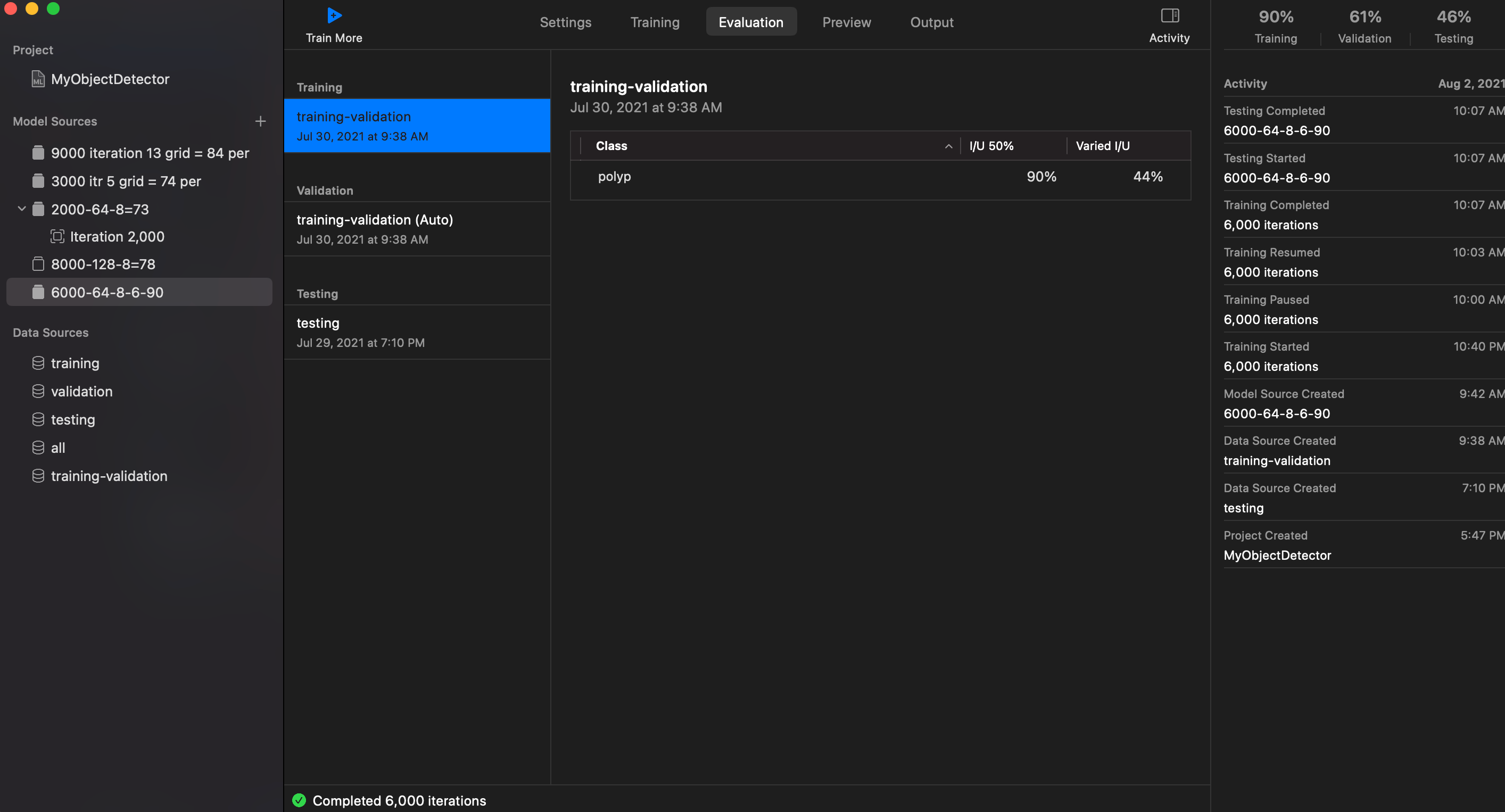
- Iteration: 6000
- Grid Size: 8 x 6 or could be 8 x 8
- Training and Valiation set: 850
- Testing set: 150
- Batch Size: 64
-
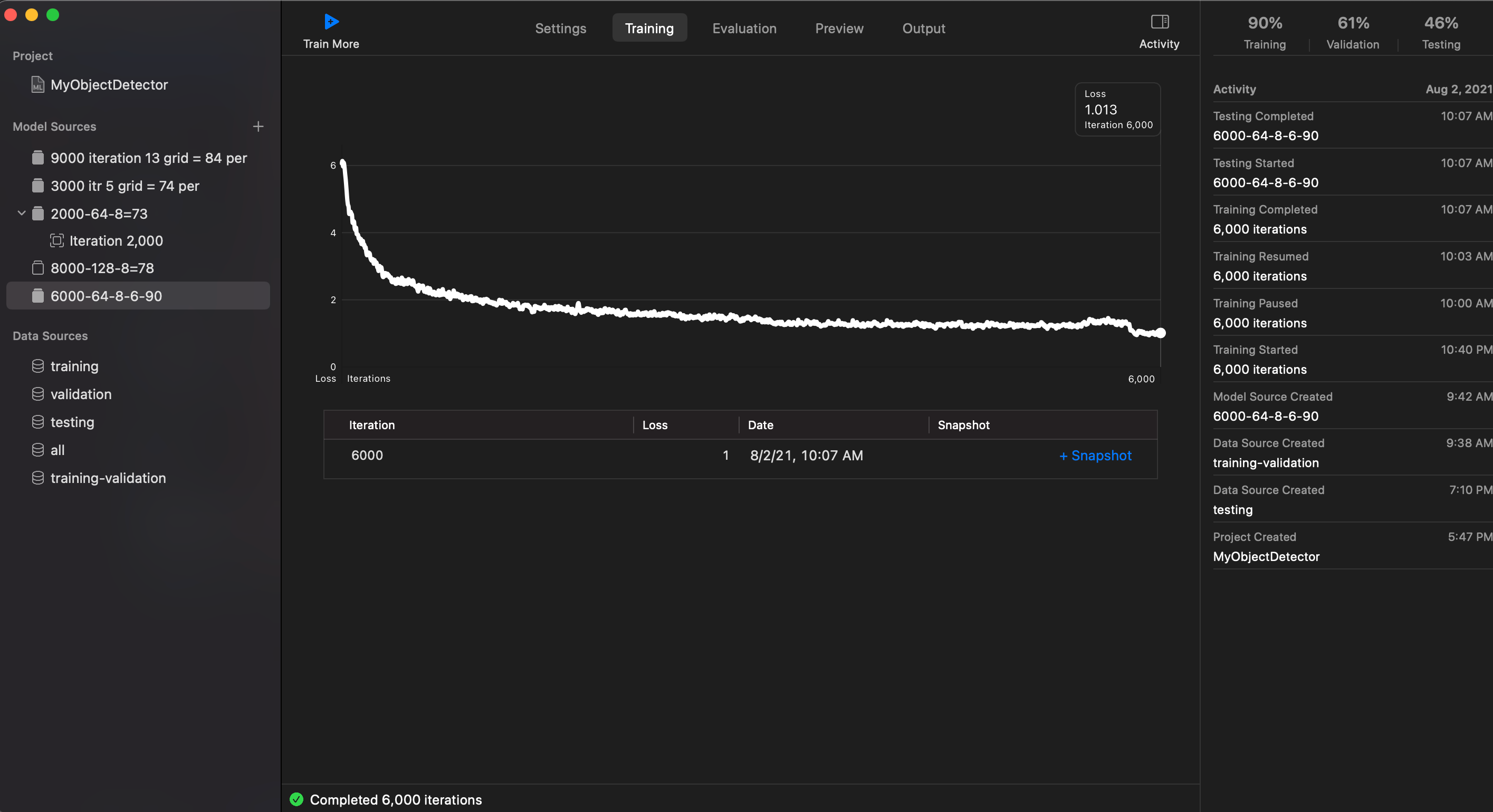
Loss: 1.013
-
I/U 50%: 90%
-
Training: 90%
-
Training: 71%
-
Training: 46%
- Download Sample Files from Hyper-Kvasir Dataset with Bounding Box https://datasets.simula.no/hyper-kvasir/
- Extract the files zip into segmented-images folder
- I wrote a little script convert.py it's located under segmented-images/convert.py. It takes hyper-vasier's bounding box json format and it converts it into Apple's CreateML annotation.json file format. It also takes 1000 images from the images folder, and split them into
- training : 700
- validation : 150
- testing: 150 it also puts training-validation folder for 850 images because I noticed that CreateML app does automatically pickup validation sets. Chose wichever you want.
- Select folders from CreateML
- Set parameter and run
- After tranining is completed, build XCode project and deploy it to your phone. I used their "Breakfast Finder" sample app. https://developer.apple.com/documentation/vision/recognizing_objects_in_live_capture
- replace ObjectDetector.mlmodel with what you outputted from CreateML project.
- Try the app on your Phone.
@misc{borgli2020, title={Hyper-Kvasir: A Comprehensive Multi-Class Image and Video Dataset for Gastrointestinal Endoscopy}, url={osf.io/mkzcq}, DOI={10.31219/osf.io/mkzcq}, publisher={OSF Preprints}, author={Borgli, Hanna and Thambawita, Vajira and Smedsrud, Pia H and Hicks, Steven and Jha, Debesh and Eskeland, Sigrun L and Randel, Kristin R and Pogorelov, Konstantin and Lux, Mathias and Nguyen, Duc T D and Johansen, Dag and Griwodz, Carsten and Stensland, H{\aa}kon K and Garcia-Ceja, Enrique and Schmidt, Peter T and Hammer, Hugo L and Riegler, Michael A and Halvorsen, P{\aa}l and de Lange, Thomas}, year={2019}, month={Dec}}