This repository contains examples of how to work with AI models for protein design.
esmfold_multimer.ipynbdemonstrates how to predict the folding of an antibody sequence using HuggingFace's ESMFold model. This model can be used to predict the folding on any protein.
antibody-affinity.ipynbdemonstrates how to load the antibody affinity dataset from TDCommons and train a neural network with PyTorch Lightning. (Currently the model makes very bad predictions.)
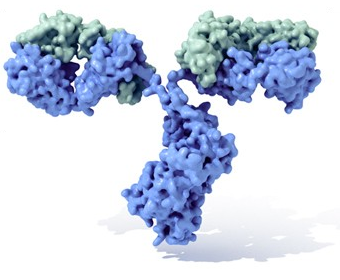
A 3D model of an Immunoglobulin molecule, showing heavy chains in blue and light chains in green. By the National Library of Medicine. Public domain.
- Install miniconda
conda env create -f environment.ymlto create the conda environment calledantibody-env.conda activate antibody-envto activate the conda environment.jupyter labto run a Jupyter Lab server. Click on the url in the output log.
Jupyter Server 2.13.0 is running at:
http://localhost:8888/lab?token=55d9dont7d55usea9fc266notgooda5d91fakeda8ae50783caedd71
- Run the notebooks.